
For each segment of a given genome, a WGA tells us where its “corresponding” segments are in other genomes. In imprecise terms, a WGA is a “correspondence” between genomes. Lastly, we lay out a number of current methodological challenges for WGA.Ģ.1 WGA as a Correspondence Between Genomes In addition to describing whole-genome aligners, we also discuss the various approaches that have been used for evaluating the alignments they produce. We then categorize the WGA methods that have been developed and describe the key computational techniques that are used within each category. We begin with a thorough definition of the problem and discuss the important downstream applications of WGAs. In this chapter, we describe the problem of WGA and the methods that address it. As orthologous positions are typically of primary interest, WGA also involves the classification of homologous relationships. In addition, a set of genomes may contain pairs of sequence positions whose evolutionary relationships can be described by any of the three major subclasses of homology: orthology, paralogy, and xenology. However, aligning whole genomes is made more complicated by the fact that genomes undergo large-scale structural changes, such as duplications and rearrangements. Like classical sequence alignment, WGA is about predicting evolutionarily related sequence positions. As each genome is sequenced, there is interest in aligning it against other available genomes in order to better understand its evolutionary history and, ultimately, the biology of its species. Whole-genome sequencing remains popular, with over 140,000 sequencing projects that are either ongoing or completed.Īlong with the ascertainment of these sequences, the problem of whole-genome alignment (WGA) has arisen. As of the writing of this chapter, there are 9071 published complete genome sequences (8380 bacterial, 281 archaeal, and 410 eukaryotic), according to the GOLD database. DNA sequencing technology has rapidly improved since that time, and as a result, we have seen an explosion in the availability of whole-genome sequences. influenzae, biologists have had access to a different scale of biological sequences, those of whole genomes. Starting in 1995 with the sequencing of the 1.8 Mb-sized genome of the bacterium H. Although limited in its scope, this type of alignment remains extremely important today, with gene-sized alignments forming the basis of most evolutionary studies. As such, classical sequence alignment (as described in Chapter 7 ) is typically focused on predicting homologous positions within two or more relatively short and colinear sequences, allowing for the edit events of substitution, insertion, and deletion.
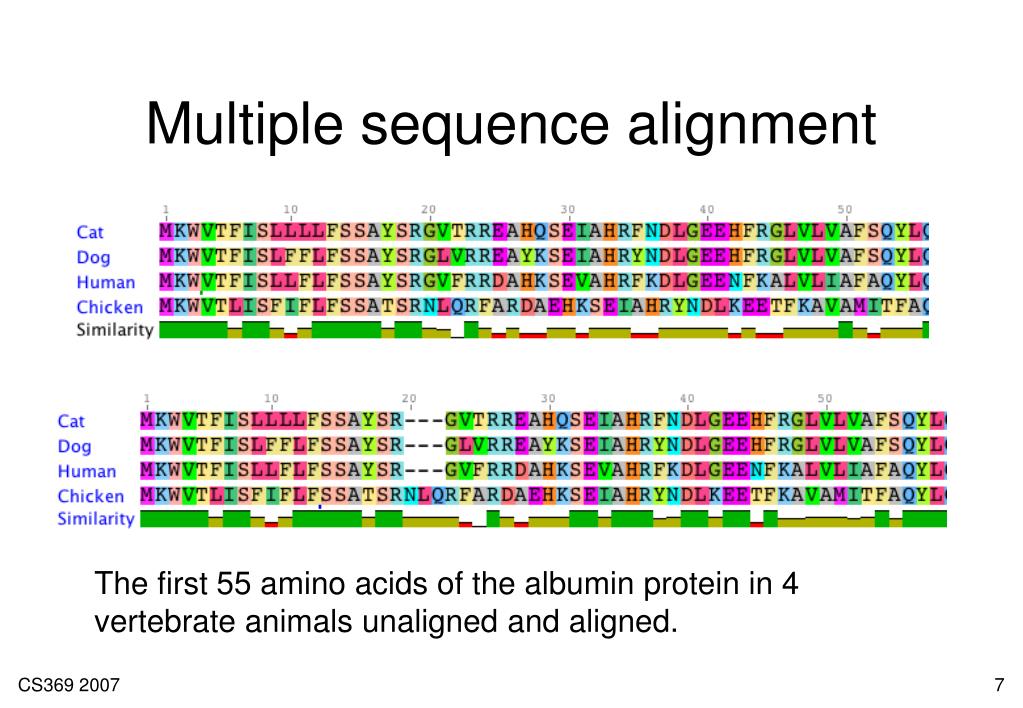
When the problem of biological sequence alignment was first described and addressed in the 1970s, sequencing technology was limited to obtaining the sequences of individual proteins or mRNAs or short genomic intervals.
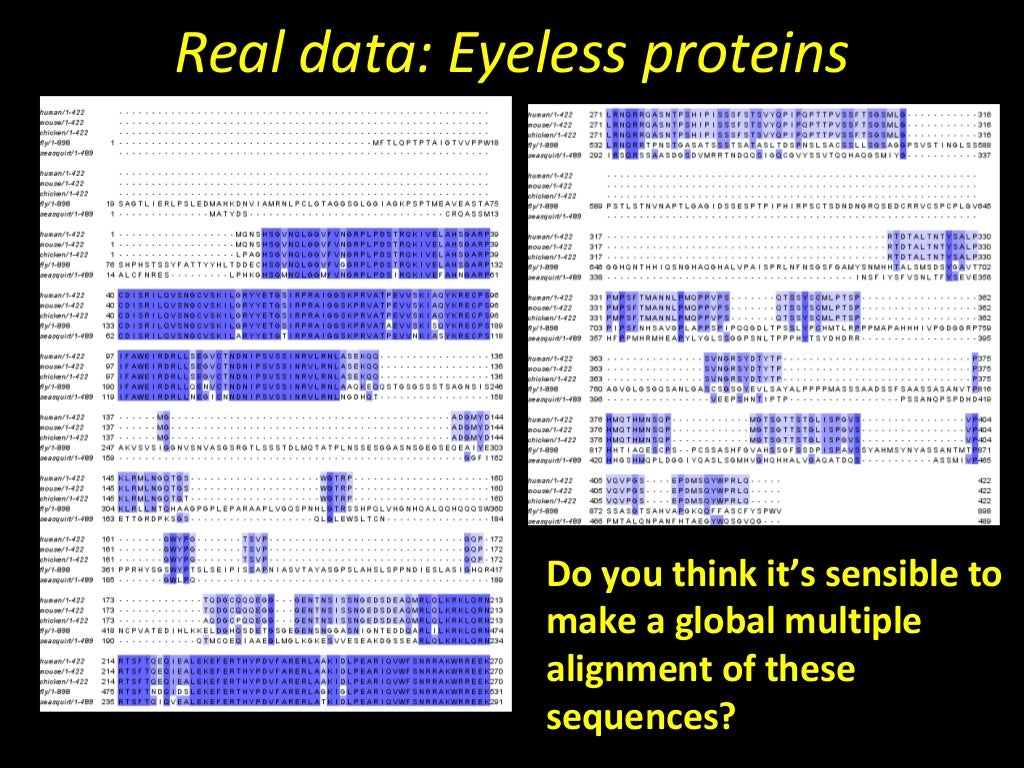
We also examine the problem of evaluating whole-genome aligners and offer a set of methodological challenges that need to be tackled in order to make most effective use of our rapidly growing databases of whole genomes. In this chapter, we discuss the meaning and significance of WGA and present an overview of the methods that address it. Despite the difficulty of this problem, numerous methods have been developed for its solution because WGAs are valuable for genome-wide analyses such as phylogenetic inference, genome annotation, and function prediction. It combines aspects of both colinear sequence alignment and gene orthology prediction and is typically more challenging to address than either of these tasks due to the size and complexity of whole genomes. Whole-genome alignment (WGA) is the prediction of evolutionary relationships at the nucleotide level between two or more genomes.


 0 kommentar(er)
0 kommentar(er)
